Table Of Content
Annealing to both exons is necessary as this ensures annealing to the exon-exon junction region but not either exon alone. Note that this option is effective only if you select "Primer must span an exon-exon junction" for "Exon junction span" option. One critical point to keep in mind is that, whether to include the stop codon or not in the primer. For example, if you want to add a tag (fluorescent protein or a short peptide etc.) to the C terminal of the protein, you need to remove the stop codon from the reverse primer to let the tag be fused during translation. If your goal is to sub-clone the gene or to express the protein without tags at the C-terminal of the gene, you will have to include the stop codon to prevent the translational fusion of vector sequences.
Polymerase Chain Reaction: Basic Protocol Plus Troubleshooting and Optimization Strategies
The reverse primer was designed from the end of the sequence that we have added to APE. The primer design is demonstrated using Dsup (Damage Suppressor) gene from tardigrade (water bear). Tardigrades are fascinating animals with extraordinary abilities to cope with extreme conditions like the vacuum of space, high tolerance for UV radiation, high and low-temperature tolerance to begin with. Dsup protein is a newly discovered gene that imparts resistance to UV radiation by coating itself to DNA.
Designing forward primer
As RT-qPCR is the current gold standard for the etiological diagnosis of SARS-CoV-2 infection, the diagnostic accuracy of this technique should be a foremost prerequisite. This review provides an overview of the most crucial components of a RT-qPCR assay-primer design (summarised in Figure Figure4).4). First, it is important to identify the reference sequence using a step-by-step process, and eventually align the sequence, if required. The gene targets include the N, E, ORF 1ab (or RdRP), and S genes. The more conserved E gene is the target for the pan-coronavirus assay, while N and RdRP genes mainly function as targets for confirmatory assays. After considering the basic rules of primer/probe design, and degenerate primers/probes against SARS-CoV can be used as a positive control; moreover, primers that account for the variation in coronavirus sequences can be developed.
Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction

Whatever the issues are, with the help of tools like APE, designing the reverse primer is just a click of a button away. Follow the steps mentioned below to get the reverse primer sequence. Since a complete alignment between a primer and its targets is desired for accurate specificity checking, the NW global alignment algorithm [13] is incorporated into Primer-BLAST to realign any regions that are not fully aligned by BLAST.
The proportion of co-infections has been underestimated due to either a lack of sensitivity or the limitations of virus detection technologies 57. As co-infections can occur, all patients that meet the suspected case definition should be tested for COVID-19 regardless of whether another respiratory pathogen is found. However, all tests shown in Table Table22 indicate no cross-reactivity with HCoV-229E, HCoV-NL63, HCoV-HKU1, HCoV-OC43, MERS-CoV, and other respiratory viruses, indicating that the specificity of the primers and probes listed in Table Table22 is high. Assays are useful only if the correct target has been identified and used in the assay design.
Note that the specificity is checked not only for the forward-reverse primer pair, but also for forward-forward as well as reverse-reverse primer pairs. The test templates are randomly selected from NCBI Refseq mRNA database and they include 52 human sequences for testing QuantPrime and 24 Arabidopsis thaliana sequences for testing PRIMEGENS (since PRIMEGENS does not support human sequences). As shown in Table 2, QuantPrime or PRIMEGENS generated primer pairs for most test cases that they deemed to be specific for input templates. However, Primer-BLAST revealed that many of these (13.4% of primer pairs from QuantPrime and 43.3% of primers pairs from PRIMEGENS) have potential unintended targets that show between one and five nucleotide mismatches.
Products and services
Designing highly multiplex PCR primer sets with Simulated Annealing Design using Dimer Likelihood Estimation ... - Nature.com
Designing highly multiplex PCR primer sets with Simulated Annealing Design using Dimer Likelihood Estimation ....
Posted: Mon, 11 Apr 2022 07:00:00 GMT [source]
Also, if you are performing a one-step reverse transcription PCR (RT-qPCR), the reverse transcriptase will use the reverse primer to prime the transcription reaction. In this scenario, a poor primer would result in both inefficient reverse transcription and inefficient amplification – a lose–lose situation. Whatever the challenge in your PCR assay, we hope that these tips for PCR primer design will be helpful to you. NEB’s long history in PCR drives us to support scientists breaking new frontiers today. We were the first company to bring Taq DNA Polymerase to the research market, the first to discover a PCR-stable, high-fidelity DNA polymerase, and the first to provide reagents for PCR performed in space. High GC primers can bind non-specifically to off target templates.
IVT Product Purification
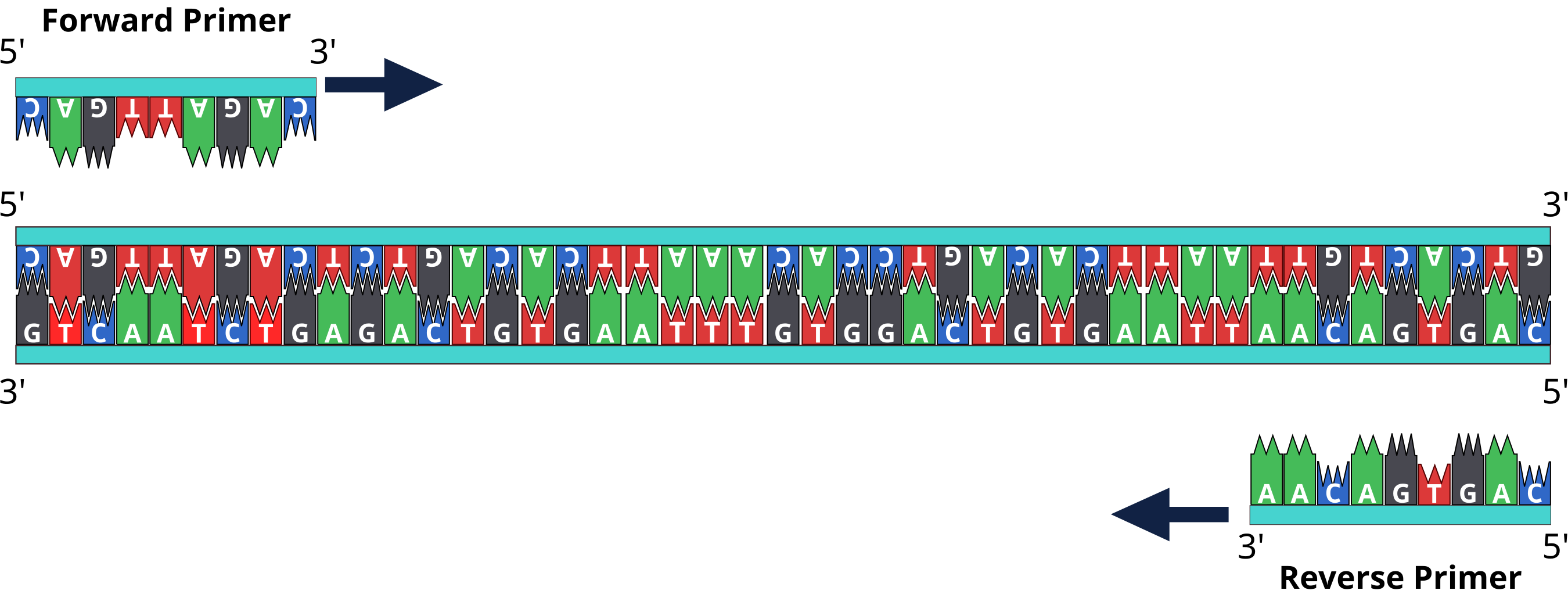
The sequence covered in blue open rectangle is used for reverse primer. Blue ribbon indicates primer sequence selected for reverse primer design. Reverse complementation contains two steps (Complementation and changing the direction) were shown. Polymerization direction is into the template stand rather outward of sequence as happens without reverse complementation. The forward and reverse primers are two different sets of short DNA fragments. Each one is complementary to a specific sequence on one of the strands on the template DNA molecule.
PCR Primer Design Tips
The design of SARS-CoV-2 primers and probes and the step-by-step analysis process to accurately detect the virus are shown in Figure Figure11. SARS-CoV-2 is an enveloped, positive-sense, single-stranded RNA virus. The genome is arranged in the order of a 5′ untranslated region (UTR)-replicase complex (open reading frame [ORF] 1ab)-structural protein [Spike(S)-Envelope(E)-Membrane (M)-Nucleocapsid (N)]-3′UTR and non-structural ORFs 4. The S-protein has a strong binding affinity to human ACE2 5, and this, along with a long incubation time before manifesting symptoms, means that SARS-CoV-2 has high transmissibility with a mortality rate of approximately 3% 6,7. This also poses considerable challenges regarding the timely treatment and effective prevention and control of COVID-19. To better understand the origin of SARS-CoV-2 and its genetic relationship with other coronaviruses, phylogenetic analyses of coronavirus sequences from various sources can be performed using the neighbour-joining method in MEGA X 8.
As a result, a large portion of test cases have at least one primer pair that has potential unintended targets (31.5% for QuantPrime and 93.3% for PRIMEGENS). Some targets have only one or two mismatches to primers generated from QuantPrime (18.5%) although this portion is much smaller for PRIMEGENS (3.4%). The specificity checking module by default uses BLAST search parameters that ensure high sensitivity such that it can detect a target that contains up to 35% mismatches to the primer sequence. The default BLAST expect value cutoff is 30,000 for the primer-only case and it is typically adjusted much higher for the template case (see below). Other highly sensitive default parameters include a word size of seven (standard BLAST uses 11), 50,000 for the maximum number of database sequences (standard BLAST uses 250) and 1 for match reward to mismatch penalty ratio (standard BLAST uses 1.5).
Previous studies have detected five different SARS-CoV-2 haplotypes, and the results indicated active genetic recombination 40. The mutation regions may reduce the accuracy of RT-qPCR detection 41. Thus, designing degenerate primers and/or probes is an effective method for addressing the challenge of virus mutations. However, no previous review has systematically described how to design primers and probes methodically and rationally in the face of a new coronavirus. Therefore, the focus of the present review is to discuss effective primer designing and improving the sensitivity and specificity of the detection process.
As reported in previous studies, primer dimers can be prevented by modifying with 2′-O-methyl bases in the penultimate base 26. The closer the probe is to the upstream primer, the higher the hydrolysis efficiency. The 5' end of the probe cannot start with a G base because it can quench the fluorophore 62.


No comments:
Post a Comment